Help |
1. What is Single-cell eQTL Hub?
Single cell approaches have already revealed that stem cell cultures contain multiple cell types that have unique transcriptional profiles. Single-cell eQTL Hub aims to comprehensively provide single cell eQTLs in different cell types. It provides a number of tools aimed at functional interpretation. The single-cell eQTL Hub includes 15 studies with 3,358 donors providing nearly 10 million cells of more than 240 different types. More than 10,000 eGenes were identified by eQTL analysis.
In single-cell eQTL Hub, users can:
Browse or search single-cell eQTLs across different cell types
Browse or search single-cell eQTLs in regions/positions
Download figures and all single-cell eQTLs results
2. How to search in Single-cell eQTL Hub?
Search by variant: you can srarch by rsID or
position, like rs1732886 or 12:66583762 (GRCh37). Please observe the
writing rules strictly.
Search by gene: you can srarch by gene symbol or
region, like IRAK3 or 12:66532659-66698402 (GRCh37). Please observe
the writing rules strictly.
3. About the search result
(1) Search by variant:
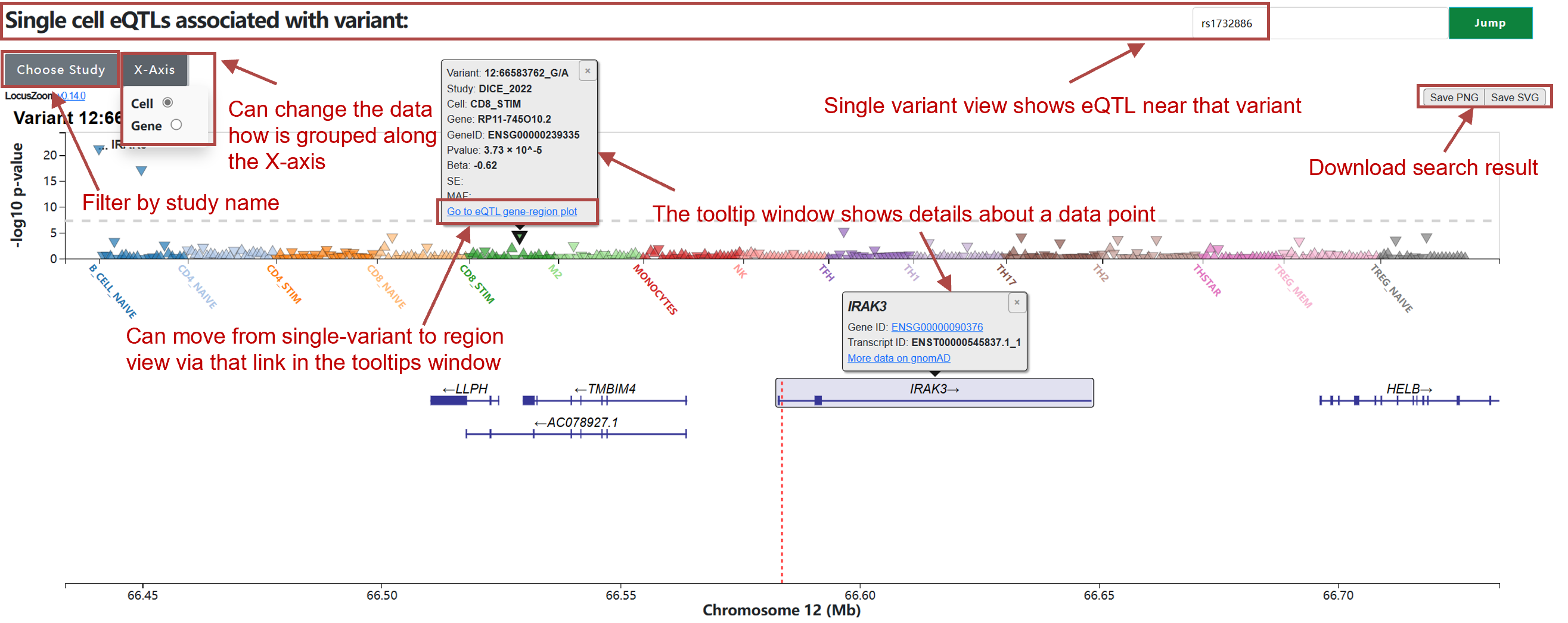

The single-cell view table is sorted by P-value.
Study: Study from PubMed, named after first author
and year of publication. See Part 5 for more details.
Beta: Effect size of SNP on gene expression.
SE: Standard error for beta values.
(2) Search by gene:
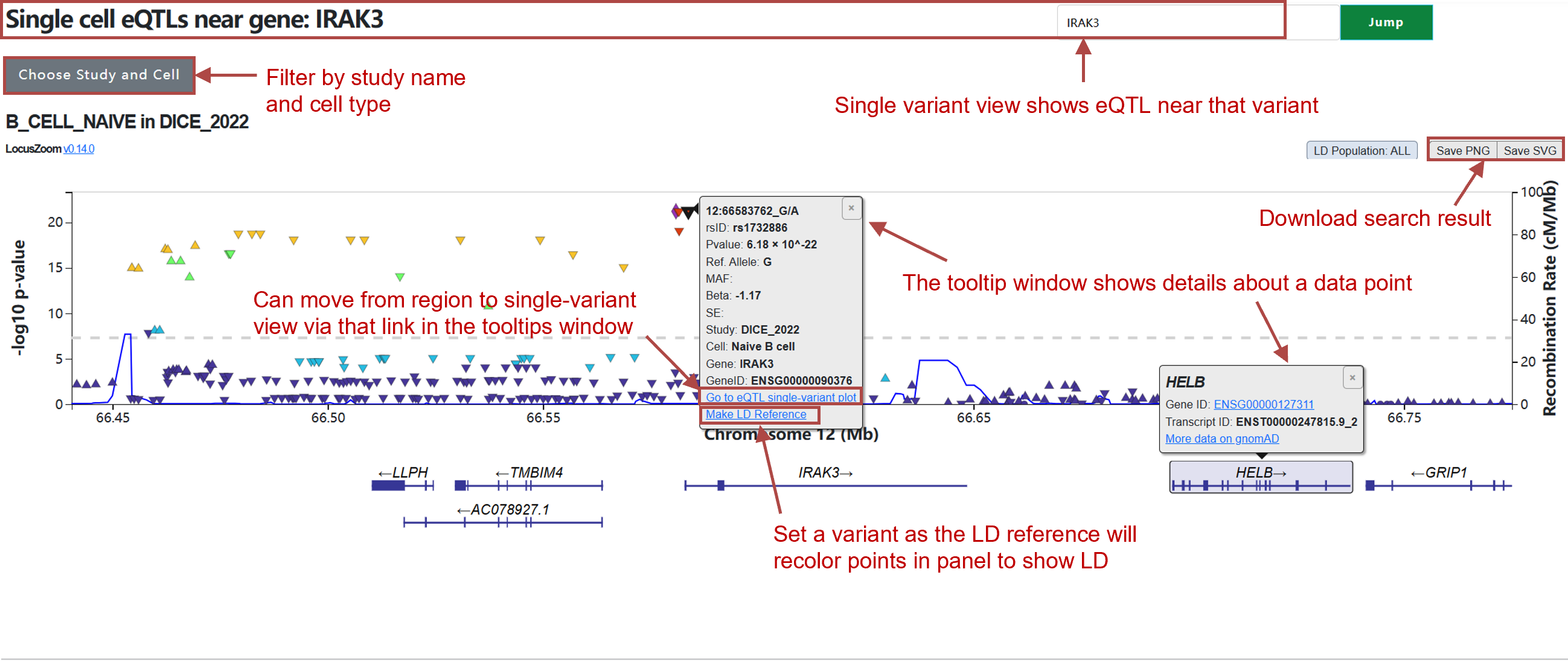
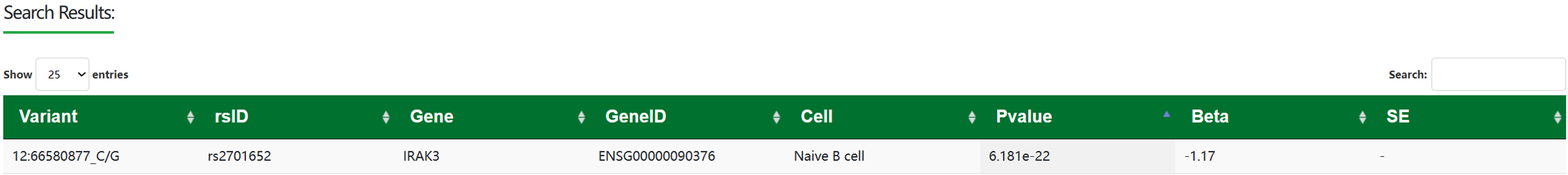
The region view table is sorted by P-value.
Variant: Contains SNP position and SNP alleles.
Beta: Effect size of SNP on gene expression.
SE: Standard error for beta values.
4. Database construction pipeline
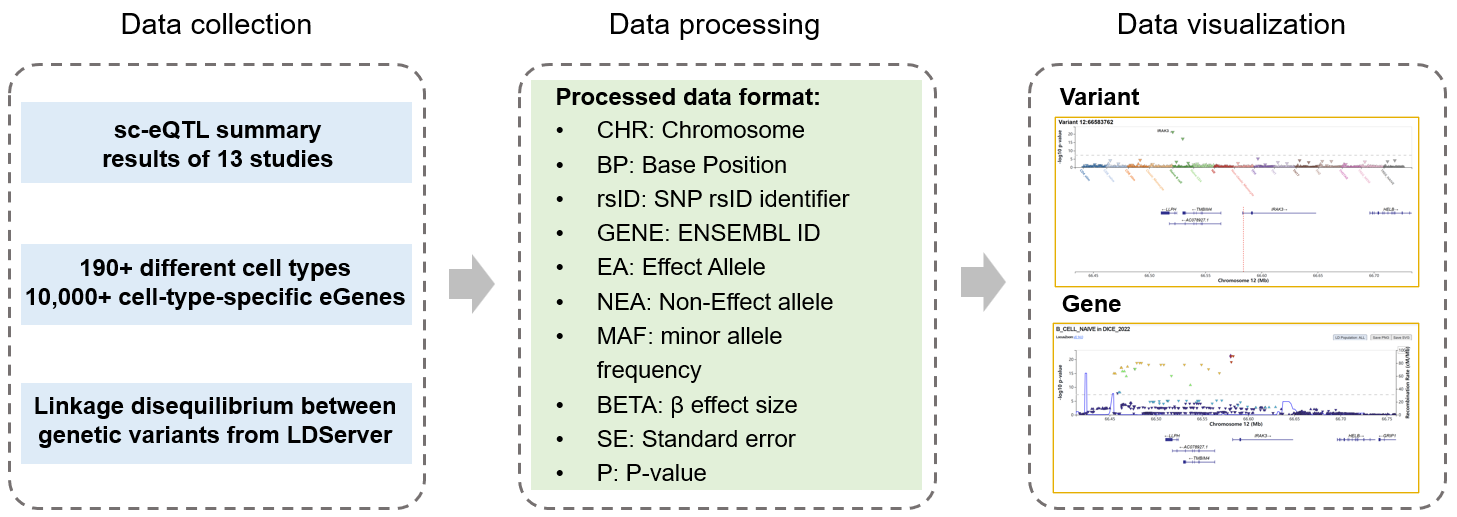
5. Data summary
We provide summary, coordination, and analysis of single-cell eQTL
data in the following articles. For more details on contributing
to the research or to download the original data, visit the
articles.
Genome positions use coordinates from human genome build GRCh37.